- GU Home
- Biowissenschaften
- Institute und Einrichtungen
- Institut MBW
- Arbeitskreise
- Abt. Müller-McNicoll
- Teaching, Courses & Methods
Teaching
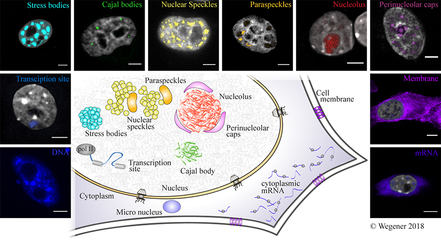
Teaching:
Bachelor Biowissenschaften
- Modul 8 (Molekularbiologie, Vorlesung, Seminar und Praktikum)
- Modul
15c (Spezialisierung Molekularbiologie, Praktikum und Seminar)
- Modul 19 (Einführung in wissenschaftliches Arbeiten, Seminar, Protokoll)
- Modul 20 (Bachelorarbeit)
Master MSc Molekulare Biowissenschaften
- Wahlmodul 9a (RNA Biologie der Eukaryoten, Vorlesung, Seminar und Praktikum)
- Modul 11 (Spezialisierung, Praktikum, Seminar, Protokoll)
- Modul 12 (Einführung in die Wissenschaftliche Arbeitstechnik, Praktikum, Seminar, Protokoll)
- Modul 13 (Masterarbeit)
Master MSc Physical Biology of Cells and Cell Interactions
- Module 3 (Advanced Cell Biology II, Lectures, Seminar)
- Module 4 (Current Concepts in Cell Biology, written proposal)
- Module 5 (Advanced Methods in Cell Biology, practical and protocol)
- Module 6 (Master thesis)
- Elective Module 33 (Cellular RNA Biologie, practical, seminar, protocol)
Courses
2018
01.06.-09.06.2018 EMBO Practical Course: iCLIP: Genomic views of protein-RNA, CRG - Centre for Genomic Regulation Barcelona, Spain
2017
26.03.-01.04.2017 EMBO Practical Course: iCLIP: Genomic views of protein-RNA, IMB Mainz, Germany
2016
07.-19.03.2016 Courses@CRG: Post-Transcriptional Gene Regulation (iCLIP & Ribosome Profiling)
2015
16.-21.11.2015 EMBO Practical Course: iCLIP - Genomic views of protein-RNA interactions
Methods
Our laboratory employs and develops quantitative and genome-wide in vivo approaches to understand the principles of eukaryotic mRNA regulation at the post-transcriptional level through RNA-binding proteins.
You can learn and apply the following methods in our lab:
- iCLIP-Seq (individual-nucleotide resolution UV cross-linking and Immuno-Precipitation followed by Deep Sequencing)
- Ribo-Seq (Ribosome footprinting followed by Deep Sequencing)
- Polysome profiling
- RNA-Seq
- Computational analysis of Deep Sequencing data
- Human and murine cell culture techniques
- Transfection of mammalian cell lines
- BAC (bacterial artificial chromosome) recombineering
- RNA interference (RNAi)
- Fluorescence microscopy
- Quantitative real-time PCR
- In-vivo purification of RNA-protein complexes
- Cloning
- Western Blotting
- Northern Blotting
Kontakt
Arbeitskreis RNA Regulation in Higher Eukaryotes
Institut für Molekulare Biowissenschaften
Prof. Michaela Müller-McNicoll, PhD
Biologicum, Campus Riedberg
Gebäudeteil B, 1.OG
Max-von-Laue-Straße 13
60438 Frankfurt am Main
T +49 69 798 - 42079
Mueller-McNicoll(at)bio.uni-frankfurt.de
Sprechzeiten:
Donnerstags, 13-14 Uhr 13-14 Uhr nur nach Vereinbarung. Mindestens eine Woche im Voraus anSekretariat:
Frau Baechle Jourdan
office-muellermcnicoll@bio.uni-frankfurt.de
- Aktuelles und Presse
- Pressemitteilungen
- Öffentliche Veranstaltungen
- Uni-Publikationen
- Aktuelles Jahrbuch
- UniReport
- Forschung Frankfurt
- Aktuelle Stellenangebote
- Frankfurter Kinder-Uni
- Internationales
- Outgoings
- Erasmus / LLP
- Goethe Welcome Centre (GWC)
- Refugees / Geflüchtete
- Erasmus +
- Sprachenzentrum oder Fremdsprachen
- Goethe Research Academy for Early Career Researchers
- Forschung
- Research Support
- Forschungsprojekte, Kooperationen, Infrastruktur
- Profilbereich Molecular & Translational Medicine
- Profilbereich Structure & Dynamics of Life
- Profilbereich Space, Time & Matter
- Profilbereich Sustainability & Biodiversity
- Profilbereich Orders & Transformations
- Profilbereich Universality & Diversity





