Research
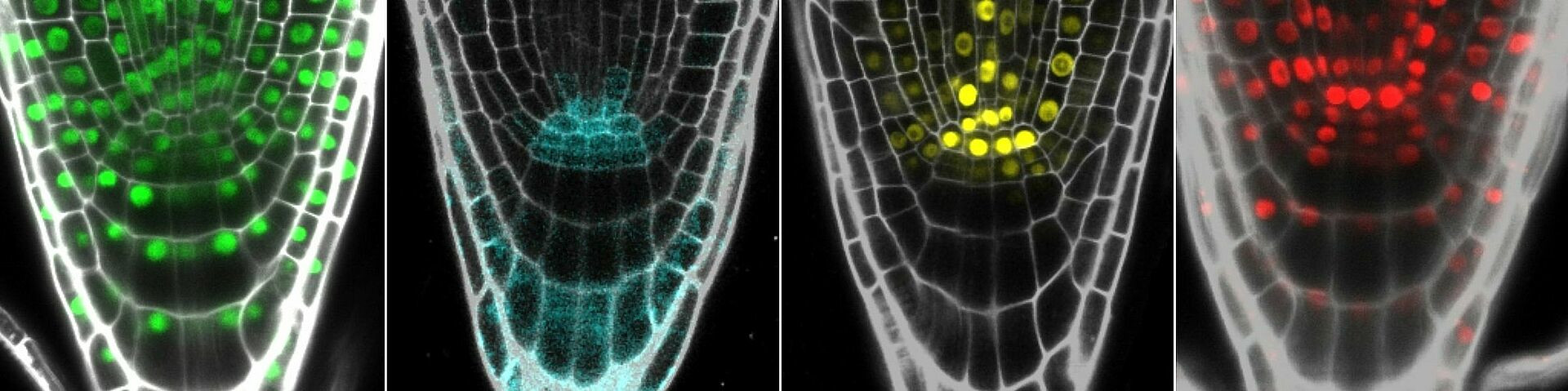
Maintenance and Function of the Barley Root Apical Meristem
Crop plants play a crucial role in food production, but climate change poses a challenge for achieving the high yields needed in view of an ever growing population. Water and nutrient availability are vital for plant growth, and roots must respond quickly to changes in soil conditions to optimize uptake. Barley has demonstrated greater tolerance to drought and salt stress compared to other crops, making it a promising organism for studying and improving root growth and development in the face of environmental challenges.
The CSCS consortium is a research unit comprised of ten groups working to understand the conserved and modified signaling pathways necessary for meristem formation and regulation in various crop species. Our project, "Maintenance and Function of the Barley Root Apical Meristem," focuses on regulating the stem cell niche in the RAM of barley.
We use two approaches to gain a deeper understanding of the underlying mechanisms. First, the unbiased approach involves single-cell RNA sequencing and spatial transcriptomics to uncover novel transcription factors and regulatory networks. Second, the biased approach investigates homologues of known transcription factors from the model plant Arabidopsis thaliana to determine whether their function is conserved across different plant species. To study the function of several transcription factors in the barley RAM, we will generate CRISPR mutants and reporter lines and employ techniques such as confocal and light sheet microscopy, as well as FRET-FLIM.
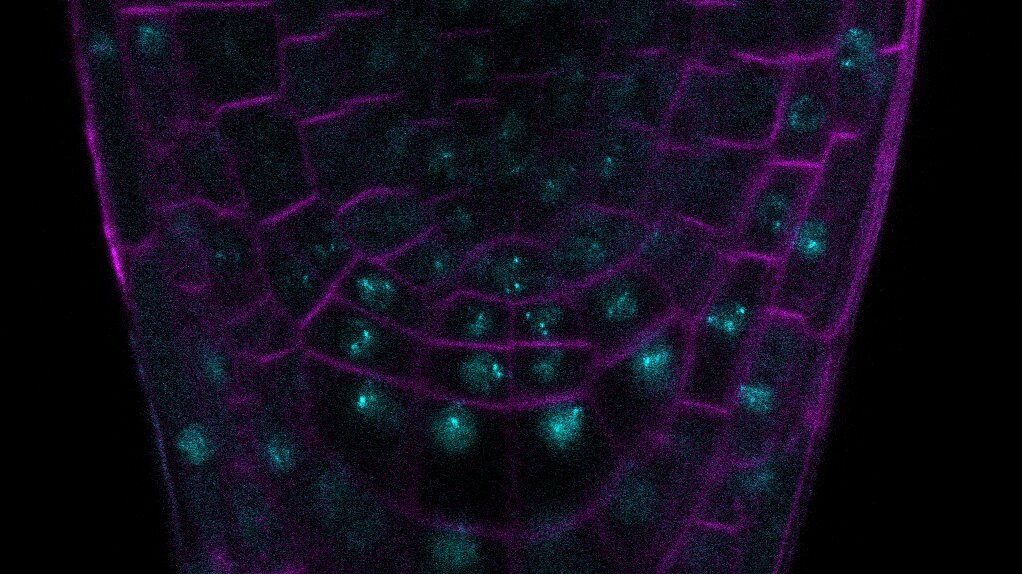
Eukaryotic cells are compartmentalized into membrane-bound organelles and membrane-less bodies, rather than being amorphous mixtures of biomolecules. The membrane-less bodies are created through liquid-liquid phase separation (LLPS), which is initiated by intrinsic and extrinsic signals. For instance, proteins with intrinsically disordered regions (IDRs), such as prion-like domains (PrDs), promote LLPS due to their flexibility, lack of a defined 3D-structure, and ability to switch conformation. Indeed, the significance of PrD-dependent LLPS in plant biology has recently been emphasized, as several studies have highlighted its role in regulating plant development and physiology.



Eukaryotic cells are compartmentalized into membrane-bound organelles and membrane-less bodies, rather than being amorphous mixtures of biomolecules. The membrane-less bodies are created through liquid-liquid phase separation (LLPS), which is initiated by intrinsic and extrinsic signals. For instance, proteins with intrinsically disordered regions (IDRs), such as prion-like domains (PrDs), promote LLPS due to their flexibility, lack of a defined 3D-structure, and ability to switch conformation. Indeed, the significance of PrD-dependent LLPS in plant biology has recently been emphasized, as several studies have highlighted its role in regulating plant development and physiology.
In the PROSPERO project, in collaboration with three French and one German partner, we are investigating the dynamics of PrDs in plant root development and their response to the environment. Transcriptional regulators that contain PrDs have recently been discovered to control root growth, development, and stress signals, and therefore act as developmental switches. By understanding how PrDs interact with the physiochemical environment, we can directly tune plant growth and stress responses.
We aim to determine the in vivo conditions under which LLPS of PrD-containing proteins occurs. For this purpose, we use various in vivo fluorescence techniques (e.g. FRAP, FRET-FLIM, fluorescence anisotropy) in simplified transient expression systems and transgenic Arabidopsis thaliana. The goal of this project is to determine the characteristics of the dynamics and stability of the condensed state in a functional biological context upon changing environmental cues.
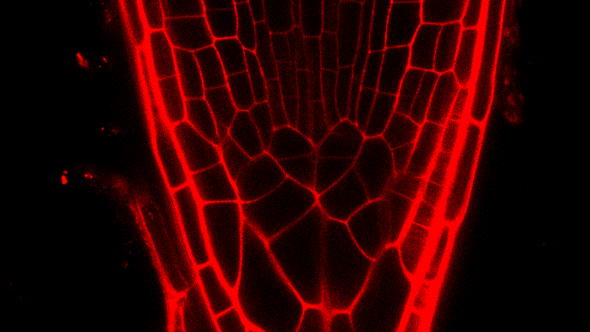
A molecular hub controlling root stem cell homeostasis
The root system is a crucial component of higher plants, providing access to water and nutrients while anchoring the plant in the soil. In the model plant Arabidopsis thaliana, the root is formed by a group of pluripotent cells, which rarely divide and form the quiescent center (QC). The QC plays a vital role in maintaining the surrounding stem cells non-cell autonomously and serves as a long-term reservoir for new cells. However, the stem cell pool needs to be replenished by coordinated QC cell divisions due to the loss of stem cells from differentiation or stress.
The balance between quiescence and stem cell replenishment is strictly regulated by several signaling pathways, including phytohormones, peptides, receptors, and transcription factors. Although many of the factors involved are well understood, the mutual regulation and interplay of these signaling pathways remain unclear. This project aims to link different regulatory pathways in the root meristem that can potentially connect hormonal, developmental, and stress-induced signals in the stem cell niche of Arabidopsis.
To achieve this, the project will use a heterologous expression system, Nicotiana benthamiana, as well as transgenic Arabidopsis lines. Traditional genetic methods and advanced microscopy techniques, such as FRET-FLIM and fluorescence anisotropy, will be combined to shed light on the unknown connections within the regulatory network controlling root stem cell maintenance in Arabidopsis.
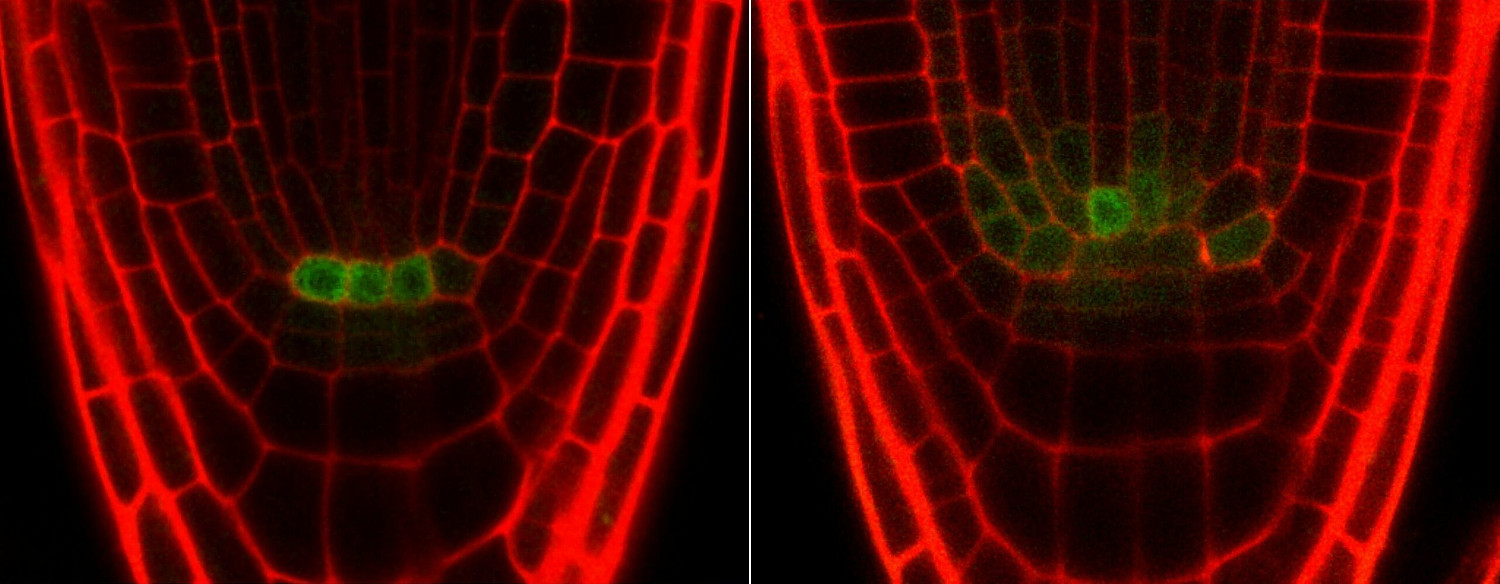

Current model of the control of QC divisions in the Arabidopsis RAM. For
the replenishment of stem cells during development or upon stresses,
different phytohormones influence the activity of TFs in order to
regulate necessary QC divisions. The QC cells are marked in red.

Maintenance and Function of the Barley Root Apical Meristem
Crop plants play a crucial role in food production, but climate change poses a challenge for achieving the high yields needed in view of an ever growing population. Water and nutrient availability are vital for plant growth, and roots must respond quickly to changes in soil conditions to optimize uptake. Barley has demonstrated greater tolerance to drought and salt stress compared to other crops, making it a promising organism for studying and improving root growth and development in the face of environmental challenges.
The CSCS consortium is a research unit comprised of ten groups working to understand the conserved and modified signaling pathways necessary for meristem formation and regulation in various crop species. Our project, "Maintenance and Function of the Barley Root Apical Meristem," focuses on regulating the stem cell niche in the RAM of barley.
We use two approaches to gain a deeper understanding of the underlying mechanisms. First, the unbiased approach involves single-cell RNA sequencing and spatial transcriptomics to uncover novel transcription factors and regulatory networks. Second, the biased approach investigates homologues of known transcription factors from the model plant Arabidopsis thaliana to determine whether their function is conserved across different plant species. To study the function of several transcription factors in the barley RAM, we will generate CRISPR mutants and reporter lines and employ techniques such as confocal and light sheet microscopy, as well as FRET-FLIM.

Eukaryotic cells are compartmentalized into membrane-bound organelles and membrane-less bodies, rather than being amorphous mixtures of biomolecules. The membrane-less bodies are created through liquid-liquid phase separation (LLPS), which is initiated by intrinsic and extrinsic signals. For instance, proteins with intrinsically disordered regions (IDRs), such as prion-like domains (PrDs), promote LLPS due to their flexibility, lack of a defined 3D-structure, and ability to switch conformation. Indeed, the significance of PrD-dependent LLPS in plant biology has recently been emphasized, as several studies have highlighted its role in regulating plant development and physiology.



Eukaryotic cells are compartmentalized into membrane-bound organelles and membrane-less bodies, rather than being amorphous mixtures of biomolecules. The membrane-less bodies are created through liquid-liquid phase separation (LLPS), which is initiated by intrinsic and extrinsic signals. For instance, proteins with intrinsically disordered regions (IDRs), such as prion-like domains (PrDs), promote LLPS due to their flexibility, lack of a defined 3D-structure, and ability to switch conformation. Indeed, the significance of PrD-dependent LLPS in plant biology has recently been emphasized, as several studies have highlighted its role in regulating plant development and physiology.
In the PROSPERO project, in collaboration with three French and one German partner, we are investigating the dynamics of PrDs in plant root development and their response to the environment. Transcriptional regulators that contain PrDs have recently been discovered to control root growth, development, and stress signals, and therefore act as developmental switches. By understanding how PrDs interact with the physiochemical environment, we can directly tune plant growth and stress responses.
We aim to determine the in vivo conditions under which LLPS of PrD-containing proteins occurs. For this purpose, we use various in vivo fluorescence techniques (e.g. FRAP, FRET-FLIM, fluorescence anisotropy) in simplified transient expression systems and transgenic Arabidopsis thaliana. The goal of this project is to determine the characteristics of the dynamics and stability of the condensed state in a functional biological context upon changing environmental cues.

A molecular hub controlling root stem cell homeostasis
The root system is a crucial component of higher plants, providing access to water and nutrients while anchoring the plant in the soil. In the model plant Arabidopsis thaliana, the root is formed by a group of pluripotent cells, which rarely divide and form the quiescent center (QC). The QC plays a vital role in maintaining the surrounding stem cells non-cell autonomously and serves as a long-term reservoir for new cells. However, the stem cell pool needs to be replenished by coordinated QC cell divisions due to the loss of stem cells from differentiation or stress.
The balance between quiescence and stem cell replenishment is strictly regulated by several signaling pathways, including phytohormones, peptides, receptors, and transcription factors. Although many of the factors involved are well understood, the mutual regulation and interplay of these signaling pathways remain unclear. This project aims to link different regulatory pathways in the root meristem that can potentially connect hormonal, developmental, and stress-induced signals in the stem cell niche of Arabidopsis.
To achieve this, the project will use a heterologous expression system, Nicotiana benthamiana, as well as transgenic Arabidopsis lines. Traditional genetic methods and advanced microscopy techniques, such as FRET-FLIM and fluorescence anisotropy, will be combined to shed light on the unknown connections within the regulatory network controlling root stem cell maintenance in Arabidopsis.


Current model of the control of QC divisions in the Arabidopsis RAM. For
the replenishment of stem cells during development or upon stresses,
different phytohormones influence the activity of TFs in order to
regulate necessary QC divisions. The QC cells are marked in red.
- Aktuelles und Presse
- Pressemitteilungen
- Öffentliche Veranstaltungen
- Uni-Publikationen
- Aktuelles Jahrbuch
- UniReport
- Forschung Frankfurt
- Aktuelle Stellenangebote
- Frankfurter Kinder-Uni
- Internationales
- Outgoings
- Erasmus / LLP
- Goethe Welcome Centre (GWC)
- Refugees / Geflüchtete
- Erasmus +
- Sprachenzentrum oder Fremdsprachen
- Goethe Research Academy for Early Career Researchers
- Forschung
- Research Support
- Forschungsprojekte, Kooperationen, Infrastruktur
- Profilbereich Molecular & Translational Medicine
- Profilbereich Structure & Dynamics of Life
- Profilbereich Space, Time & Matter
- Profilbereich Sustainability & Biodiversity
- Profilbereich Orders & Transformations
- Profilbereich Universality & Diversity





