AK RNA Regulation in Higher Eukaryotes
The role of RBPs and subnuclear architecture in spatio-temporal gene regulation under stress
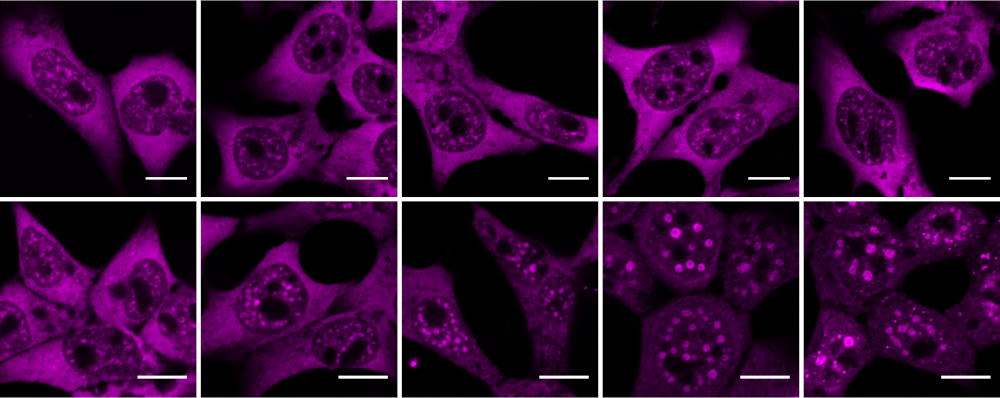
Gene expression includes all steps of the life cycle of an mRNA, starting with transcription and splicing of the pre-mRNA at the chromatin, followed by 3'end processing and polyadenylation, export through nuclear pores and finally by translation and degradation in the cytoplasm. All these steps are regulated and quality checked by RNA-binding proteins (RBPs) that form large complexes with mRNAs called mRNPs. When exposed to stress, cells rapidly and globally change their gene expression program. Reprogramming occurs in the context of a crowded and compartmentalized nucleus that contains various architectural elements such as the nuclear lamina, nuclear pore complexes (NPCs) and chromatin domains, and membraneless organelles (MLOs) such as nucleoli, Cajal bodies, nuclear speckles and paraspeckles.
MLOs are very dynamic structures that assemble either through liquid-liquid phase separation (LLPS) or through the scaffolding by architectural RNAs (arcRNAs) into more meshwork-like condensates. Most MLOs cannot be biochemically purified from cells, thus their exact composition remains unknown. However, some of them appear to change their composition as well as their shape under stress, or even assemble de novo. MLOs appear to be featureless in cryo-electron micrographs, probably due to a very high local concentration of their components and to their lack of regular structure, which limits the contrast within MLOs and with to the surrounding nucleoplasm.
Sorting, concentrating or sequestrating specific RNA and protein components from the surrounding nucleoplasm allows a fast, reversible and effective way to adjust global gene expression. Moreover, MLOs with distinct physicochemical properties would allow to organize the long pre-mRNAs and long non-coding RNAs in space to prevent entangling and to facilitate packaging, processing and quality control of RNAs by locally enriched processing enzymes.
We want to understand how MLOs assemble or remodel under stress and whether they adopt specific ultrastructures. We investigate how MLOs organize transcripts and RBPs in space, how they globally reprogram gene expression and how they contribute to nuclear RNA surveillance.
As model MLOs we focus on paraspeckles and nuclear speckles and their abundant component SR proteins. SR proteins (SRSF1-SRSF12), control mRNA metabolism from transcription in the nucleus to translation and decay in the cytoplasm. SR proteins are known for their essential roles in the selection of splice sites and the regulation of alternative splicing (AS), but individual SR protein family members also perform non-canonical functions, which are much less understood.
We want to understand the role of SR proteins in paraspeckle and nuclear speckle dynamics and their crosstalk, discover novel functions of these proteins and decipher the underlying mechanisms, and assess their contribution to the regulation of mRNA metabolism in response to stress or disease.
To answer these questions, we develop new methods and quantitative assays and combine them with quantitative light microscopy, super-resolution microscopy, structural biology, reporter gene assays, protein biochemistry, mass spectrometry, transcriptomics, ribonomics and computational analyses. We use different mammalian cell culture systems and CRISPR/Cas9 genome editing and perturb cellular homoeostasis through hypoxia, oxidative stress, splicing inhibition or differentiation.
- Aktuelles und Presse
- Pressemitteilungen
- Öffentliche Veranstaltungen
- Uni-Publikationen
- Aktuelles Jahrbuch
- UniReport
- Forschung Frankfurt
- Aktuelle Stellenangebote
- Frankfurter Kinder-Uni
- Internationales
- Outgoings
- Erasmus / LLP
- Goethe Welcome Centre (GWC)
- Refugees / Geflüchtete
- Erasmus +
- Sprachenzentrum oder Fremdsprachen
- Goethe Research Academy for Early Career Researchers
- Forschung
- Research Support
- Forschungsprojekte, Kooperationen, Infrastruktur
- Profilbereich Molecular & Translational Medicine
- Profilbereich Structure & Dynamics of Life
- Profilbereich Space, Time & Matter
- Profilbereich Sustainability & Biodiversity
- Profilbereich Orders & Transformations
- Profilbereich Universality & Diversity




